library(tidyverse)
library(tidymodels)Introduction
Strokes Gained is an interesting method for evaluating golfers, co-created by Columbia Business School professor Mark Broadie.
Strokes Gained co-creator @markbroadie illustrates the statistic using Justin Thomas' final-round eagle at the 2021 #THEPLAYERS. 🔎 pic.twitter.com/LJ6qnp3ooE
— Golf Channel (@GolfChannel) March 8, 2023
From my understanding, Strokes Gained is similar to Expected Points Added (EPA) in football - golfers are evaluated against an “expected” number of strokes remaining after each shot. This “expected” value is based off a predictive model trained on historical data1. In this post, I’ll build an expected strokes remaining model using PGA ShotLink data, which can later be used to estimate strokes gained. The model will use features in the shot-level data to predict how many strokes remaining the golfer has before the shot.
Data Preparation
First, I’ll load the cleaned up data from my previous post.
shot_level <- readRDS("../02_shotlink_explore/shot_level.rds")
cut_colors <- readRDS("../02_shotlink_explore/cut_colors.rds")Before I start building a model, I want to gain a better understanding of how penalties, drops, and provisionals are handled in the data. The shot_type_s_p_d column has this information (S = Shot, P = Penalty, D = Drop, Pr = Provisional).
#Filter to holes where player's had at least 1 penalty/drop/provisional
shot_level %>%
mutate(is_p_d = ifelse(shot_type_s_p_d == 'S',0,1)) %>%
group_by(player,
round,
hole) %>%
mutate(p_d_count = sum(is_p_d)) %>%
filter(p_d_count > 0) %>%
ungroup() %>%
arrange(player,
round,
hole,
shot) %>%
select(player,
round,
hole,
shot,
type = shot_type_s_p_d,
strokes = num_of_strokes,
yards_out = yards_out_before_shot,
to_location) %>%
as.data.frame() %>%
head(14) player round hole shot type strokes yards_out to_location
1 2206 2 18 1 S 1 238.00000000 Rough
2 2206 2 18 2 D 0 13.16666667 Rough
3 2206 2 18 3 S 1 15.11111111 Green
4 2206 2 18 4 S 1 1.16666667 Hole
5 6527 4 6 1 S 1 222.00000000 Water
6 6527 4 6 2 P 1 20.25000000 Other
7 6527 4 6 3 D 0 20.25000000 Other
8 6527 4 6 4 S 1 70.11111111 Green
9 6527 4 6 5 S 1 0.02777778 Hole
10 6567 1 6 1 S 1 215.00000000 Water
11 6567 1 6 2 P 1 13.27777778 Other
12 6567 1 6 3 D 0 13.27777778 Other
13 6567 1 6 4 S 1 73.36111111 Green
14 6567 1 6 5 S 1 3.66666667 HoleReviewing the sample above, it looks like the yards_out_before_shot column (which will probably be the best predictor of strokes remaining) is a little misleading for penalty drops. For example, it looks like Player 6527 went in the water off the tee on the 6th Hole (Day 4), and had to take a penalty drop. The yards_out_before_shot value on the penalty and the drop is 20.25, but 70.11 on the first shot after the drop. This might be because ShotLink is measuring to where the ball landed in the water, but Player 6527 had to drop where they entered the penalty area. For my model, I’ll filter down to actual shots, where shot_type_s_p_d = "S". I’ll also add a shot_id so it will be easier to join back to the original data set later.
shot_level <- shot_level %>%
arrange(player,
round,
hole,
shot) %>%
mutate(shot_id = row_number())
shots <- shot_level %>%
filter(shot_type_s_p_d == 'S')Linear Model
Now I’ll take a look at how distance from the hole correlates to strokes remaining.
shots %>%
ggplot(mapping = aes(x = yards_out_before_shot,
y = strokes_remaining_before_shot)) +
geom_jitter(width = 0,
height = 0.1,
alpha = 0.25) +
geom_smooth(method = loess,
se = FALSE) +
scale_y_continuous(breaks = 1:10)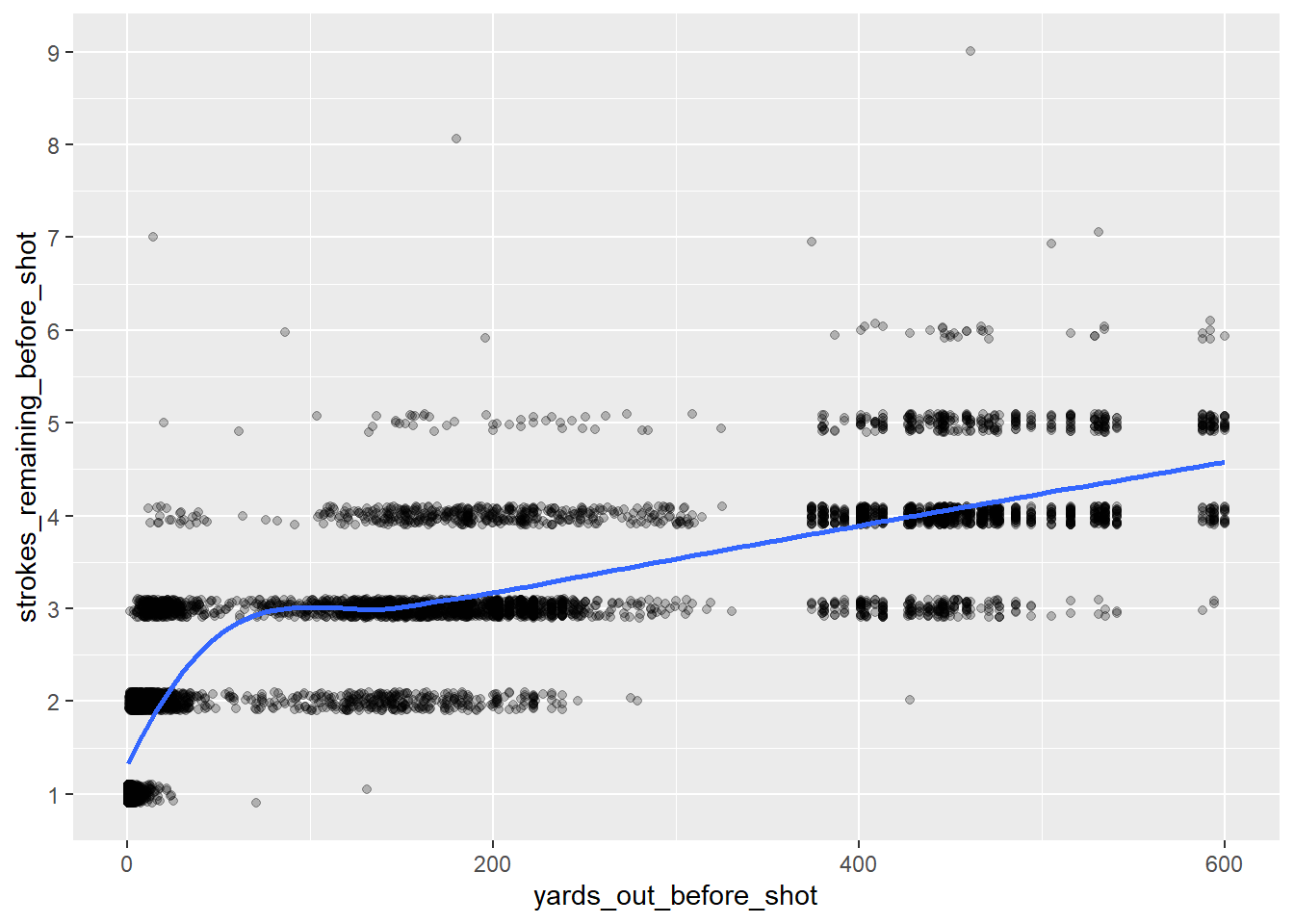
#Calculate R-Squared
cor(shots$yards_out_before_shot,
shots$strokes_remaining_before_shot)^2[1] 0.682005While there is a certainly a strong correlation between these numbers, the relationship is not quite linear. A log transformation should clean this up.
shots <- shots %>%
mutate(log_yards_out_before_shot = log(yards_out_before_shot+1))
shots %>%
ggplot(mapping = aes(x = log_yards_out_before_shot,
y = strokes_remaining_before_shot)) +
geom_jitter(width = 0,
height = 0.1,
alpha = 0.1) +
geom_smooth(method = loess,
se = FALSE) +
scale_y_continuous(breaks = 1:10)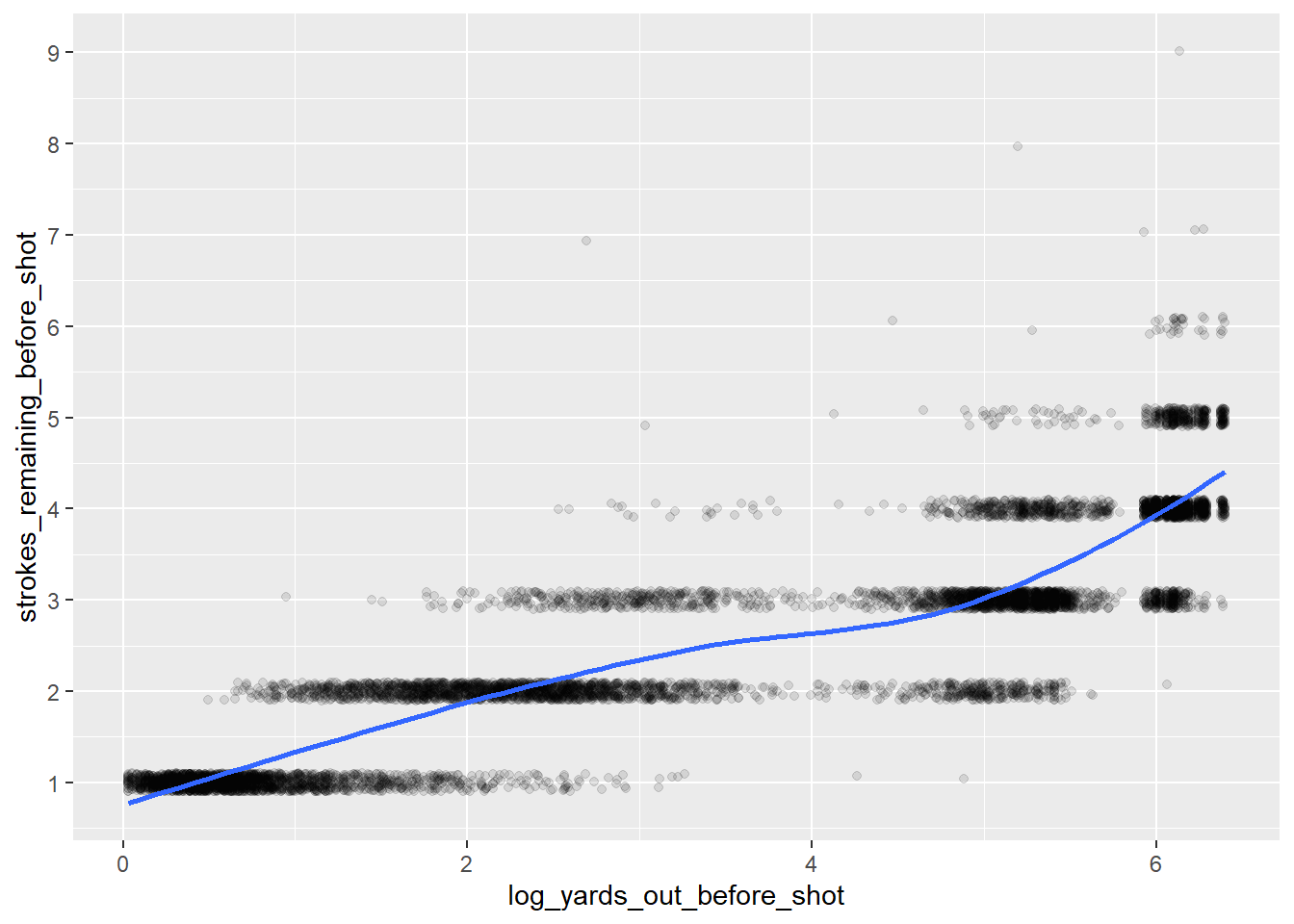
#Calculate R-Squared
cor(shots$log_yards_out_before_shot,
shots$strokes_remaining_before_shot)^2[1] 0.7876248The log transformation improves the R2 value, but it can probably be improved even further with a nonlinear model. To test this, I’ll use cross-validation to evaluate out-of-sample performance. Ideally, I’d like to use some type of time-series data splitting here to avoid any possible data leakage issues2, but I’ll use a simpler method in this post. I’ll split the 30 golfers into 10 groups of 3, and hold out one of these groups in the cross-validation process.
player_cv_groups <- shots %>%
select(player) %>%
unique() %>%
arrange(player) %>%
mutate(cv_group = cut_number(x = player,
n = 10,
labels = FALSE))
shots <- shots %>%
inner_join(player_cv_groups,
by = "player")
shots %>%
group_by(cv_group) %>%
summarize(golfers = n_distinct(player),
shots = n())# A tibble: 10 × 3
cv_group golfers shots
<int> <int> <int>
1 1 3 834
2 2 3 840
3 3 3 832
4 4 3 834
5 5 3 820
6 6 3 835
7 7 3 839
8 8 3 845
9 9 3 834
10 10 3 835folds <- group_vfold_cv(data = shots,
group = cv_group)Now, using these folds, I’ll find a performance baseline using the linear model above.
just_log_yards_recipe <- recipe(formula = strokes_remaining_before_shot ~
log_yards_out_before_shot,
data = shots)
lm_mod <- linear_reg(mode = "regression",
engine = "lm")
lm_workflow <- workflow() %>%
add_recipe(just_log_yards_recipe) %>%
add_model(lm_mod)
lm_rs <- fit_resamples(object = lm_workflow,
resamples = folds,
control = control_resamples(save_pred = T))
lm_rs %>%
collect_metrics() %>%
select(.metric,
mean) %>%
as.data.frame() .metric mean
1 rmse 0.5569516
2 rsq 0.7900181As expected, the mean hold-out performance is very similar to the linear model fit on all the data.
XGBoost Model
Next I’ll try a gradient boosted tree model using the XGBoost library. I love XGBoost. It trains and tunes relatively quickly, and you don’t usually need to worry about other tedious pre-processing steps like centering, scaling, and imputing. Since XGBoost is nonlinear, I wont need to use the log transformation, and can switch back to just using yards_out_before_shot.
just_yards_recipe <- recipe(formula = strokes_remaining_before_shot ~
yards_out_before_shot,
data = shots)
xgb_mod <- boost_tree(mode = "regression",
engine = "xgboost")
xgb_workflow <- workflow() %>%
add_recipe(just_yards_recipe) %>%
add_model(xgb_mod)
xgb_rs <- fit_resamples(object = xgb_workflow,
resamples = folds,
control = control_resamples(save_pred = T))
xgb_rs %>%
collect_metrics() %>%
select(.metric,
mean) %>%
as.data.frame() .metric mean
1 rmse 0.5236481
2 rsq 0.8142939The XGBoost model improves performance without any parameter tuning, so hopefully I can get more out of it. First, I’ll look at the out-of-sample predictions,
rs_preds <- xgb_rs %>%
collect_predictions() %>%
select(row_id = .row,
fold = id,
pred = .pred)
preds_join <- shots %>%
as.data.frame() %>%
mutate(row_id = row_number()) %>%
inner_join(rs_preds,
by = "row_id")
preds_join %>%
ggplot(mapping = aes(x = yards_out_before_shot,
y = pred,
color = fold)) +
geom_point()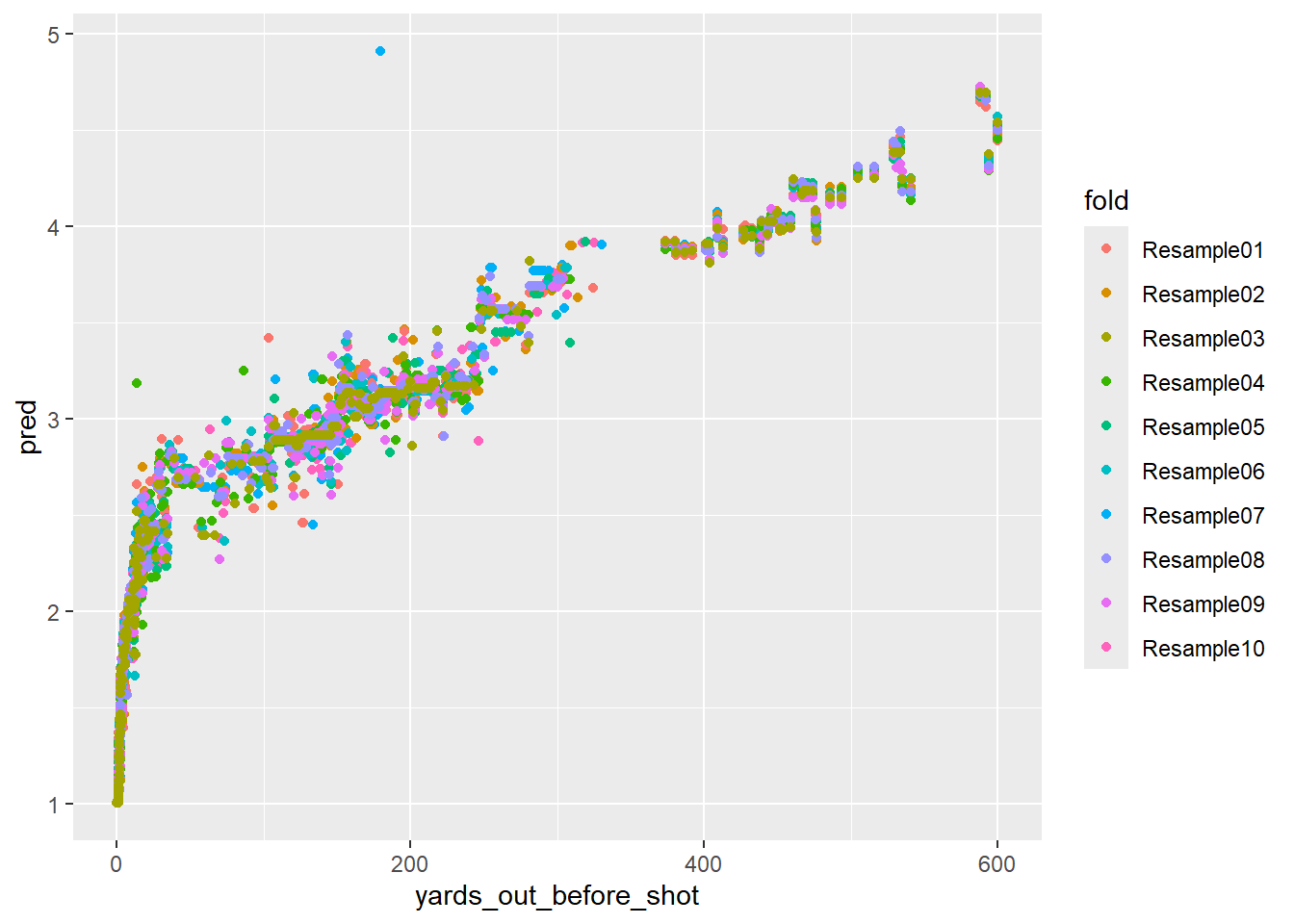
The fits look good, but the nonlinear warts are showing. This model only uses one feature, yards_out_before_shot, but the predictions vary significantly at the same distance. For example, looking at the shots around 200 yards out, the predictions vary from 2.8ish to 3.5ish. This will cause confusion when we start attributing strokes gained to specific shots.
Monotonic Constraints
Luckily, there is a fix - XGBoost has the ability to enforce monotonic constraints, meaning I can force predicted strokes remaining to increase as yards out increases. I’ll retrain my model.
xgb_mod <- boost_tree(mode = "regression") %>%
set_engine(engine = "xgboost",
monotone_constraints = c(1))
xgb_workflow <- workflow() %>%
add_recipe(just_yards_recipe) %>%
add_model(xgb_mod)
xgb_rs <- fit_resamples(object = xgb_workflow,
resamples = folds,
control = control_resamples(save_pred = T))
xgb_rs %>%
collect_metrics() %>%
select(.metric,
mean) %>%
as.data.frame() .metric mean
1 rmse 0.5201534
2 rsq 0.8168157The model performance actually improved slightly with the constraints, which is encouraging. Now I’ll look at the out-of-sample predictions again.
rs_preds <- xgb_rs %>%
collect_predictions() %>%
select(row_id = .row,
fold = id,
pred = .pred)
preds_join <- shots %>%
as.data.frame() %>%
mutate(row_id = row_number()) %>%
inner_join(rs_preds,
by = "row_id")
preds_join %>%
ggplot(mapping = aes(x = yards_out_before_shot,
y = pred,
color = fold)) +
geom_point()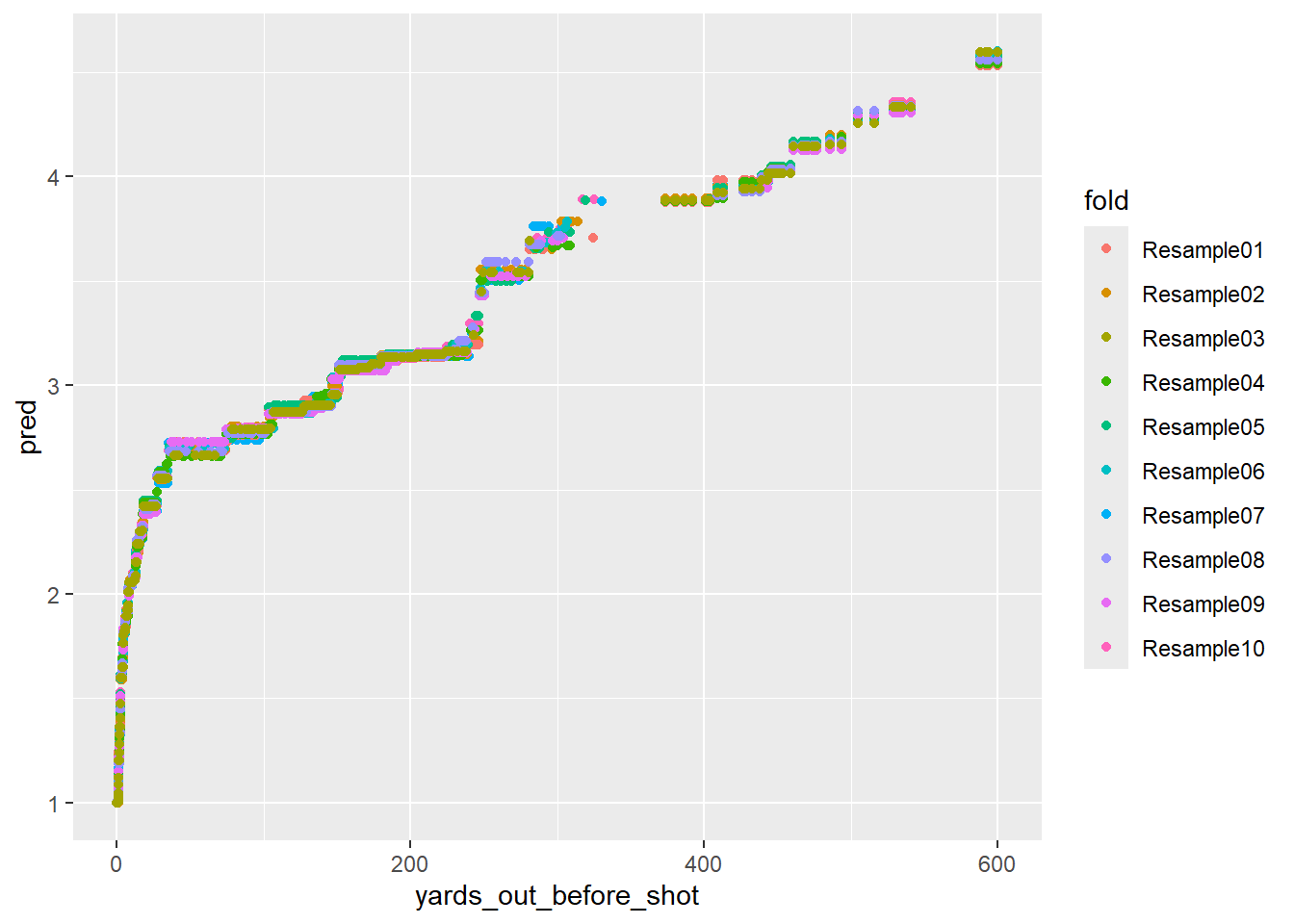
Much better! Another interesting finding is that there is a “jump” in the predictions around 260 yards. The predicted strokes remaining jump from 3.2ish to 3.5ish. This could be because it’s near the range where PGA Tour players can take more aggressive shots at the green to reduce their score. This is a good demonstration of the value of nonlinear models.
Ball Location Features
Next, I’d like to add some more features. I’ll start with the ball location (fairway, green, etc.). I cleaned up ball location data in my last post, but those were the to_location columns. Here I need the from_location columns because I am using strokes_remaining_before_shot and yards_out_before_shot.
shots %>%
group_by(from_location_scorer,
from_location_laser) %>%
summarize(rows = n(),
.groups = "keep") %>%
arrange(desc(rows)) %>%
as.data.frame() from_location_scorer from_location_laser rows
1 Green Unknown 3546
2 Tee Box 2162
3 Fairway Left Fairway 550
4 Fairway Right Fairway 533
5 Primary Rough Left Rough 415
6 Primary Rough Right Rough 375
7 Intermediate Rough Right Intermediate 144
8 Intermediate Rough Left Intermediate 112
9 Fairway Bunker Unknown 92
10 Green Side Bunker Right Green Side Bunker 77
11 Green Side Bunker Right Front Green Side Bunker 66
12 Green Side Bunker Front Left Green Side Bunker 59
13 Green Side Bunker Left Green Side Bunker 47
14 Green Side Bunker Front Center Green Side Bunker 34
15 Fringe Left Fairway 28
16 Other Unknown 20
17 Primary Rough Unknown 20
18 Fringe Right Fairway 18
19 Native Area Left Rough 13
20 Native Area Unknown 13
21 Native Area Right Rough 11
22 Green Side Bunker Left Rear Green Side Bunker 3
23 Other Left Rough 3
24 Green 2
25 Other Right Rough 2
26 Water Unknown 2
27 Green Side Bunker Right Rear Green Side Bunker 1shots <- shots %>%
mutate(from_location = case_when(from_location_scorer == 'Green'
~ 'Green',
from_location_scorer == 'Tee Box'
~ 'Tee Box',
from_location_scorer %in% c('Fairway',
'Fringe')
~ 'Fairway',
from_location_scorer %in% c('Primary Rough',
'Intermediate Rough')
~ 'Rough',
from_location_scorer %in% c('Fairway Bunker',
'Green Side Bunker')
~ 'Bunker',
TRUE
~ 'Other')) %>%
mutate(from_location = factor(from_location,
ordered = T,
levels = c("Green",
"Fairway",
"Tee Box",
"Rough",
"Bunker",
"Other")))
shots %>%
filter(player == 1810,
round == 1,
hole %in% 1:3) %>%
select(hole,
shot,
to_location,
from_location) %>%
arrange(hole,
shot) %>%
as.data.frame() hole shot to_location from_location
1 1 1 Rough Tee Box
2 1 2 Green Rough
3 1 3 Green Green
4 1 4 Hole Green
5 2 1 Green Tee Box
6 2 2 Hole Green
7 3 1 Fairway Tee Box
8 3 2 Green Fairway
9 3 3 Green Green
10 3 4 Hole GreenNow I’ll train a new XGBoost model with this feature. Since I’ve updated the data set, I’ll need to recreate the fold index and the recipe. I’ll use step_dummy() to convert from_location from a factor to binary terms for each location type.
folds <- group_vfold_cv(data = shots,
group = cv_group)
with_location_recipe <- recipe(formula = strokes_remaining_before_shot ~
yards_out_before_shot +
from_location,
data = shots) %>%
step_dummy(from_location)
xgb_mod <- boost_tree() %>%
set_mode("regression") %>%
set_engine("xgboost",
monotone_constraints = c(1)) %>%
translate()
xgb_workflow <- workflow() %>%
add_recipe(with_location_recipe) %>%
add_model(xgb_mod)
xgb_rs <- fit_resamples(object = xgb_workflow,
resamples = folds,
control = control_resamples(save_pred = T))
xgb_rs %>%
collect_metrics() %>%
select(.metric,
mean) %>%
as.data.frame() .metric mean
1 rmse 0.5124754
2 rsq 0.8221943Not much of an improvement, but the model got a little better.
rs_preds <- xgb_rs %>%
collect_predictions() %>%
select(row_id = .row,
fold = id,
pred = .pred)
preds_join <- shots %>%
as.data.frame() %>%
mutate(row_id = row_number()) %>%
inner_join(rs_preds,
by = "row_id")
preds_join %>%
ggplot(mapping = aes(x = yards_out_before_shot,
y = pred,
color = from_location)) +
geom_point() +
scale_color_manual(values = cut_colors) +
labs(title = "Out-Of-Sample Predicted Strokes Remaining")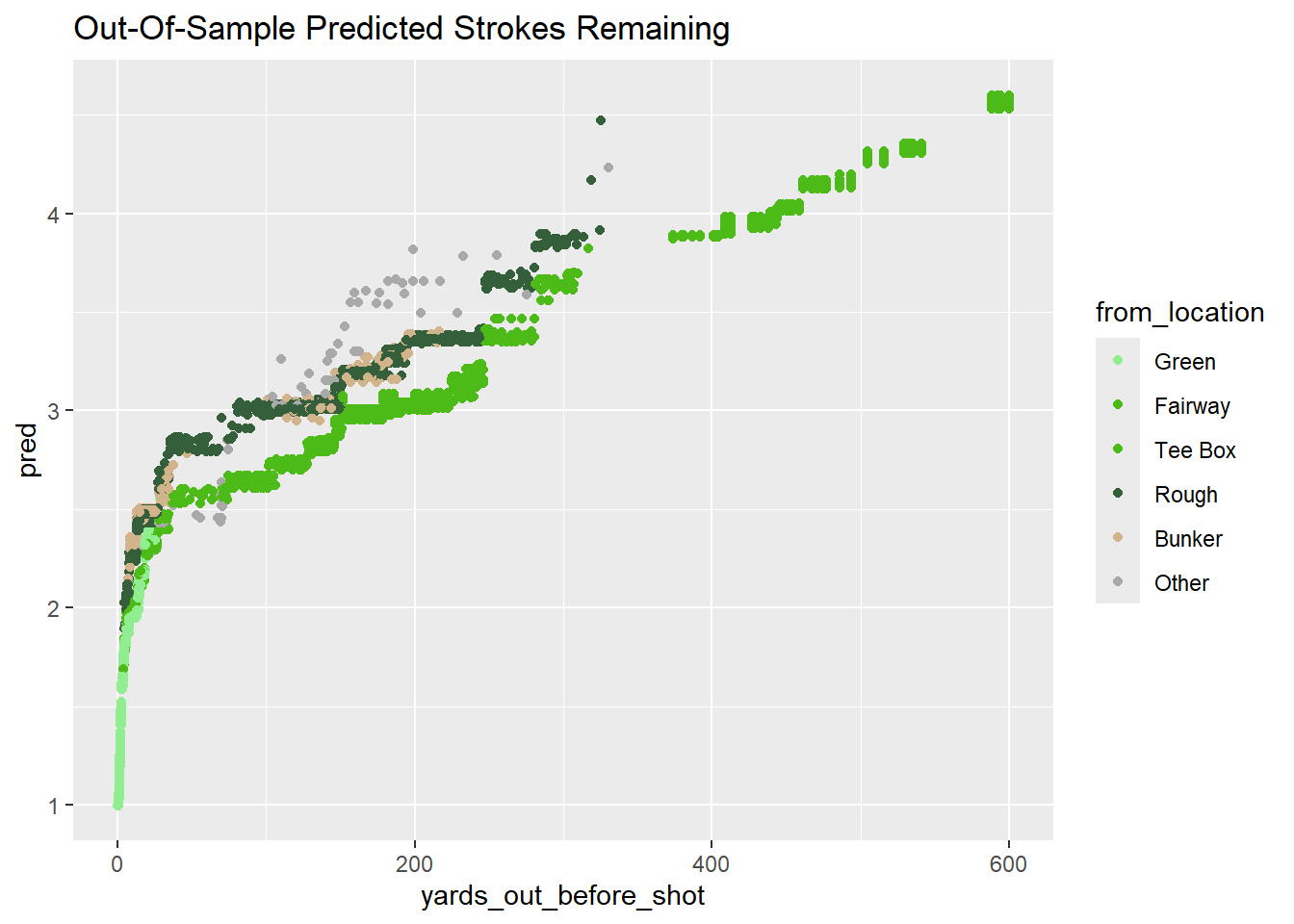
These new predictions make sense. Shots from the rough/bunker are harder and thus the predicted strokes remaining are higher. For example, at 100 yards, being in the rough (vs. fairway) increases expected strokes from 2.7ish to 3.0ish.
Adding these features increased the prediction “noise” at each distance - similar to the results of the unconstrained XGBoost model. This stems from the variation in how each sub-model handles the new features, and it goes away if I filter down to a single hold out set (Fold 1).
preds_join %>%
filter(fold == 'Resample01') %>%
ggplot(mapping = aes(x = yards_out_before_shot,
y = pred,
color = from_location)) +
geom_point() +
scale_color_manual(values = cut_colors) +
labs(title = "Out-Of-Sample Predicted Strokes Remaining",
subtitle = "Fold 1")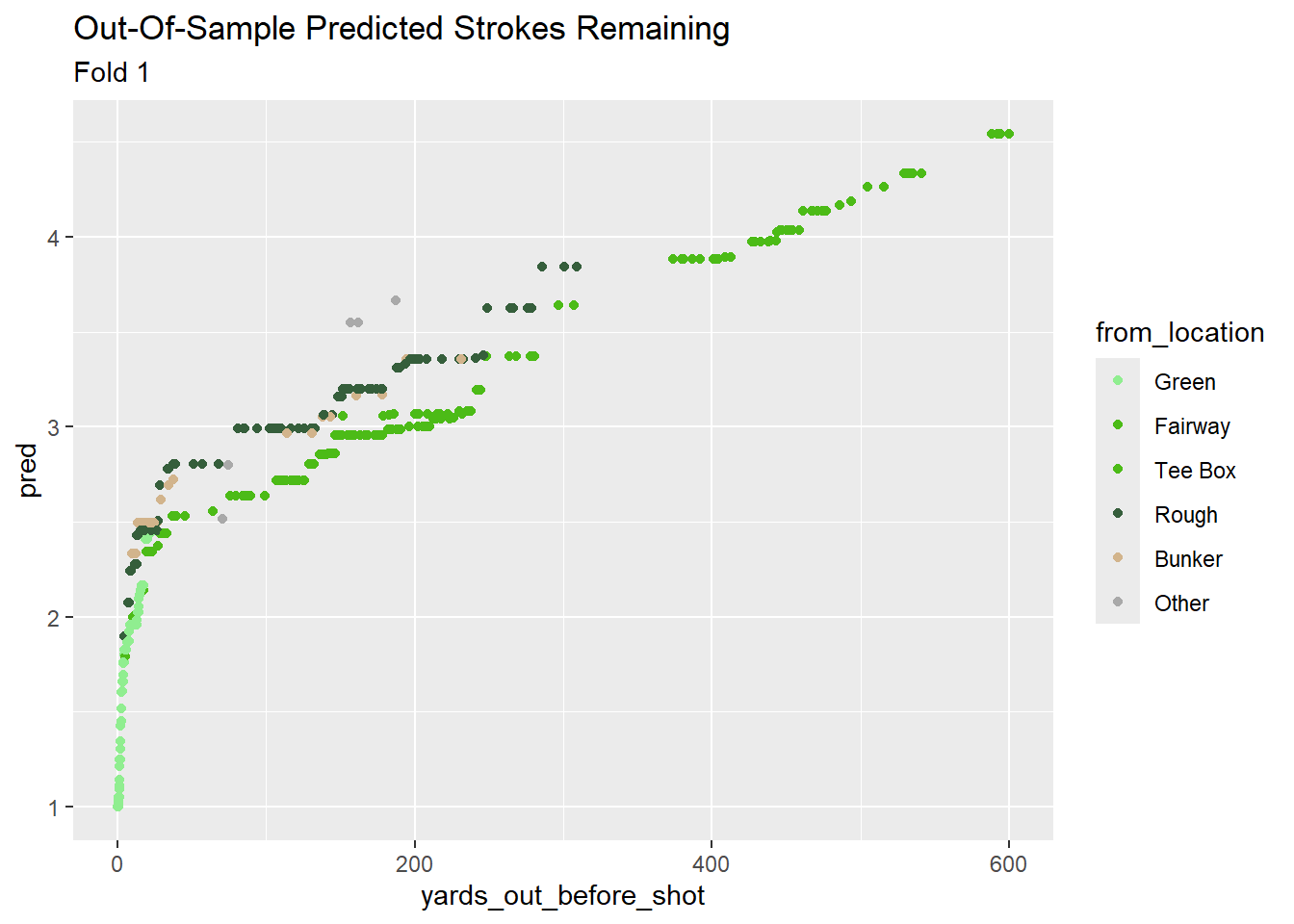
I could probably improve model performance with some parameter tuning, but I’ll stop here for now.
Footnotes
These methods have a major shortcoming in that they attribute the entire residual to a single golfer (or the teams involved). This could probably be corrected with a hierarchical/mixed model, but I’ll save that for a future post.↩︎
If I wanted to use this model to predict strokes remaining for future shot’s, I would want to make sure I’m not using future shots when training/evaluating my model.↩︎